Lectures
▾
Lecture 1
Lecture 2
Lecture 3
Lecture 4
Lecture 5
Lecture 6
Lecture 7
Lecture 8
Lecture 9
Lecture 10
Lecture 11
Skills Lab
▾
Skills lab 1
Skills lab 2
Skills lab 3
Skills lab 4
Skills lab 5
Skills lab 6
Skills lab 7
Skills lab 8
Skills lab 9
Skills lab 10
Practicals
▾
Practical 1
Practical 2
Practical 3
Practical 4
Practical 5
Practical 6
Practical 7
Practical 8
Practical 9
Practical 10
Practical 11
Tutorials
▾
Tutorial 0
Tutorial 1
Tutorial 2
Tutorial 3
Tutorial 4
Tutorial 5
Tutorial 6
Tutorial 7
Tutorial 8
Tutorial 9
Tutorial 10
More
▾
Documents
Visualisations
About
This is the 2022 version of the Analysing Data website. For the current incarnation,
click here
.
PDF
class: middle, inverse, title-slide # Fundamentals of Statistical Testing ### Dr Milan Valášek ### 24 January 2022 --- ## Housekeeping - Welcome from Jennifer and Milan - Familiarise yourself with Canvas and AnD website - Come to practicals and bring laptops - Help desk sessions from this week (sign up on Canvas) - More info in this week's practical -- <br> - **It won't be easy so do put in the hours!** --- ## Overview - Recap on distributions - More about the normal distribution - Sampling - Sampling distribution - Standard error - Central Limit Theorem --- ## Objectives After this lecture you will understand - that there exist mathematical functions that describe different distributions - what makes the normal distribution normal and what are its properties - how random fluctuations affect sampling and parameter estimates - the function of the sampling distribution and the standard error - the Central Limit Theorem .center[**With this knowledge you'll build a solid foundation for understanding all the statistics we will be learning in this programme!**] --- ## It's all Greek to me! - `\(\mu\)` is the *population* mean - `\(\bar{x}\)` is the *sample* mean - `\(\hat{\mu}\)` is the **estimate** of the *population* mean - Same with *SD*: `\(\sigma\)`, `\(s\)`, and `\(\hat{\sigma}\)` - Greek is for populations, Latin is for samples, hat is for population estimates --- ## Recap on distributions - Numerically speaking, the number of observations per each value of a variable - Which values occur more often and which less often - The shape formed by the bars of a bar chart/histogram .codePanel[ ```r df <- tibble(eye_col = sample(c("Brown", "Blue", "Green", "Gray"), 555, replace = T, prob = c(.55, .39, .04, .02)), age = rnorm(length(eye_col), 20, .65)) p1 <- df %>% ggplot(aes(x = eye_col)) + geom_bar(fill = c("skyblue4", "chocolate4", "slategray", "olivedrab"), colour=NA) + labs(x = "Eye colour", y = "Count") p2 <- df %>% ggplot(aes(x = age)) + geom_histogram() + stat_density(aes(y = ..density.. * 80), geom = "line", color = theme_col, lwd = 1) + labs(x = "Age (years)", y = "Count") plot_grid(p1, p2) ``` <!-- -->] --- ## Known distributions - Some shapes are "algebraically tractable", <i>e.g.</i>, there is a maths formula to draw the line - We can use them for statistics .codePanel[ ```r df <- tibble(x = seq(0, 10, length.out = 100), norm = dnorm(scale(x), sd = .5), chi = dchisq(x, df = 2) * 2, t = dt(scale(x), 5, .5), beta = (dbeta(x / 10, .5, .5) / 4) - .15) cols <- c("#E69F00", "#56B4E9", "#009E73", "#CC79A7") df %>% ggplot(aes(x = x)) + geom_line(aes(y = norm), color = cols[1], lwd = 1) + geom_line(aes(y = chi), color = cols[2], lwd = 1) + geom_line(aes(y = t), color = cols[3], lwd = 1) + geom_line(aes(y = beta), color = cols[4], lwd = 1) + labs(x = "x", y = "Density") ``` <img src="data:image/png;base64,#slides_files/figure-html/unnamed-chunk-2-1.png" height="350" />] --- ## The normal distribution - AKA Gaussian distribution, The bell curve - The one you **need to** understand - Symmetrical and bell-shaped - _Not every_ symmetrical bell-shaped distribution is normal! - It's also about the proportions - The normal distribution has fixed proportions and is a function of two parameters, `\(\mu\)` (mean) and `\(\sigma\)` (or _SD_; standard deviation) --- ## The normal distribution - Peak/centre of the distribution is its mean (also mode and median) - Changing mean (**centring**) shifts the curve left/right - *SD* determines steepness of the curve (small `\(\sigma\)` = steep curve) - Changing *SD* is also known as **scaling** <iframe id="norm-dist-viz" class="viz app" src="https://and.netlify.app/viz/norm_dist" data-external="1" style="height:485px;-webkit-transform: scale(.9) translateY(-70px);"></iframe> --- ### Area below the normal curve - No matter the particular shape of the given normal distribution, the proportions with respect to *SD* are the same - **∼68.2%** of the area below the curve is within **±1 *SD*** from the mean - **∼95.4%** of the area below the curve is within **±2 *SD*** from the mean - **∼99.7%** of the area below the curve is within **±3 *SD*** from the mean - We can calculate the proportion of the area with respect to any two points .codePanel[ ```r quantiles <- tibble(x1 = -(1:3), x2 = 1:3, y = c(.21, .12, .03)) tibble(x = seq(-4, 4, by = .1), y = dnorm(x, 0, 1)) %>% ggplot(aes(x, y)) + geom_density(stat = "identity", color = default_col) + geom_density(data = ~ subset(.x, x <= quantiles$x1[3]), stat = "identity", color = default_col, fill = default_col) + geom_density(data = ~ subset(.x, x >= quantiles$x2[3]), stat = "identity", color = default_col, fill = default_col) + geom_density(data = ~ subset(.x, x >= quantiles$x1[2] & x <= quantiles$x2[2]), stat = "identity", color = NA, fill = second_col) + geom_density(data = ~ subset(.x, x >= quantiles$x1[1] & x <= quantiles$x2[1]), stat = "identity", color = NA, fill = theme_col) + geom_segment(data = quantiles, aes(x = x1, xend = x2, y = y, yend = y), arrow = arrow(length = unit(0.2, "cm"), angle = 15, type = "closed", ends = "both"), color = c(bg_col, default_col, default_col)) + geom_line(data = tibble(x = rep(c(-1, 1), 2), y = rep(c(.12, .03), each = 2)), aes(x, y, group = y), color = bg_col) + geom_line(data = tibble(x = rep(c(-(2:3), 2:3), each = 2), y = as.vector(rbind(0, rep(quantiles$y[-1] + .005, 2)))), aes(x, y, group = x), lty = 2, color = default_col) + annotate("text", x = rep(0, 3), y = quantiles$y + .05, label = c("68.2%", "95.4%", "99.7%"), color = bg_col) + labs(x = "z-score", y = "Density") + scale_x_continuous(breaks = -4:4) ``` <!-- -->] --- ### Area below the normal curve - Say we want to know the number of *SD*s from the mean beyond which lie the outer 5% of the distribution .codePanel[ ```r quantiles <- qnorm(.025) * c(-1, 1) tibble(x = sort(c(quantiles, seq(-4, 4, by = .1))), y = dnorm(x, 0, 1)) %>% ggplot(aes(x, y)) + geom_line(color = default_col) + geom_density(data = ~ subset(.x, x >= quantiles[1]), stat = "identity", color = NA, fill = second_col) + geom_density(data = ~ subset(.x, x <= quantiles[2]), stat = "identity", color = NA, fill = second_col) + geom_line(data = tibble(x = rep(quantiles, each = 2), y = c(0, .15, 0, .15)), aes(x, y, group = x), lty = 2, color = default_col) + geom_segment(data = tibble(x = quantiles, xend = c(4, -4), y = c(.15, .15), yend = c(.15, .15)), aes(x = x, xend = xend, y = y, yend = yend), arrow = arrow(length = unit(0.2, "cm"), angle = 15, type = "closed"), color = default_col) + annotate("text", x = c(-2.8, 2.8), y = .18, label = ("2.5%"), color = default_col) + labs(x = "z-score", y = "Density") ``` <img src="data:image/png;base64,#slides_files/figure-html/unnamed-chunk-4-1.png" height="250" />] .smallCode[ ```r qnorm(p = .025, mean = 0, sd = 1) # lower cut-off ``` ``` ## [1] -1.959964 ``` ```r qnorm(p = .975, mean = 0, sd = 1) # upper cut-off ``` ``` ## [1] 1.959964 ``` ] --- ### Critical values - If *SD* is known, we can calculate the cut-off point (critical value) for **any proportion** of normally distributed data .smallCode[ ```r qnorm(p = .005, mean = 0, sd = 1) # lowest .5% ``` ``` ## [1] -2.575829 ``` ```r qnorm(p = .995, mean = 0, sd = 1) # highest .5% ``` ``` ## [1] 2.575829 ``` ```r # most extreme 40% / bulk 60% qnorm(p = .2, mean = 0, sd = 1) ``` ``` ## [1] -0.8416212 ``` ```r qnorm(p = .8, mean = 0, sd = 1) ``` ``` ## [1] 0.8416212 ``` ] - Other known distributions have different cut-offs but the principle is the same --- exclude: ![:live] .pollEv[ <iframe src="https://embed.polleverywhere.com/discourses/IXL29dEI4tgTYG0zH1Sef?controls=none&short_poll=true" width="800px" height="600px"></iframe> ] --- ## Sampling from distributions - Collecting data on a variable = randomly sampling from distribution - The underlying distribution is often assumed to be normal - Some variables might come from other distributions - Reaction times: *log-normal* distribution - Number of annual casualties due to horse kicks: *Poisson* distribution - Passes/fails on an exam: *binomial* distribution --- ## Sampling from distributions - Samples from the same population differ from one another ```r # draw a sample of 10 from a normally distributed # population with mean 100 and sd 15 rnorm(n = 6, mean = 100, sd = 15) ``` ``` ## [1] 101.61958 80.95560 89.62080 96.04378 106.40106 86.21514 ``` ```r # repeat rnorm(6, 100, 15) ``` ``` ## [1] 80.31573 107.63193 85.82520 99.95288 93.55956 74.73945 ``` --- ## Sampling from distributions - Statistics ( `\(\bar{x}\)`, `\(s\)`, <i>etc.</i>) of two samples will be different - **Sample** statistic (<i>e.g.</i>, `\(\bar{x}\)`) will likely differ from the **population** parameter (<i>e.g.</i>, `\(\mu\)`) ```r sample1 <- rnorm(50, 100, 15) sample2 <- rnorm(50, 100, 15) mean(sample1) ``` ``` ## [1] 98.56429 ``` ```r mean(sample2) ``` ``` ## [1] 105.4175 ``` --- ## Sampling from distributions - Statistics ( `\(\bar{x}\)`, `\(s\)`, <i>etc.</i>) of two samples will be different - **Sample** statistic (<i>e.g.</i>, `\(\bar{x}\)`) will likely differ from the **population** parameter (<i>e.g.</i>, `\(\mu\)`) .codePanel[ ```r p1 <- ggplot(NULL, aes(x = sample1)) + geom_histogram(bins = 15) + geom_vline(xintercept = mean(sample1), color = second_col, lwd = 1) + labs(x = "x", y = "Frequency") + ylim(0, 8) p2 <- ggplot(NULL, aes(x = sample2)) + geom_histogram(bins = 15) + geom_vline(xintercept = mean(sample2), color = second_col, lwd = 1) + labs(x = "x", y = "") + ylim(0, 8) plot_grid(p1, p2) ``` <!-- -->] --- ## Sampling distribution - If we took all possible samples of a given size (say *N = 50*) from the population and each time calculated `\(\bar{x}\)`, the means would have their own distribution - This is the **sampling distribution** of the mean - Approximately **normal** - Centred around the **true population mean**, `\(\mu\)` - Every statistic has its own sampling distribution (not all normal though!) --- ## Sampling distribution ```r x_bar <- replicate(100000, mean(rnorm(50, 100, 15))) mean(x_bar) ``` ``` ## [1] 99.99395 ``` .codePanel[ ```r ggplot(NULL, aes(x_bar)) + geom_histogram(bins = 51) + geom_vline(xintercept = mean(x_bar), col = second_col, lwd = 1) + labs(x = "Sample mean", y = "Frequency") ``` <!-- -->] --- exclude: ![:live] .pollEv[ <iframe src="https://embed.polleverywhere.com/discourses/IXL29dEI4tgTYG0zH1Sef?controls=none&short_poll=true" width="800px" height="600px"></iframe> ] --- ## Standard error - Standard deviation of the sampling distribution is the **standard error** ```r sd(x_bar) ``` ``` ## [1] 2.122072 ``` - Sampling distribution of the mean is *approximately normal*: *~68.2% of means of samples* of size 50 from this population will be *within ±2.12 of the true mean* --- ## Standard error - Standard error can be **estimated** from any of the samples <m>$$\widehat{SE} = \frac{SD}{\sqrt{N}}$$</m> .smallCode[ ```r samp <- rnorm(50, 100, 15) sd(samp)/sqrt(length(samp)) ``` ``` ## [1] 1.872102 ``` ```r # underestimate compared to actual SE sd(x_bar) ``` ``` ## [1] 2.122072 ``` ] - If ~68.2% of sample means lie within ±1.87, then there's a ~68.2% probability that `\(\bar{x}\)` will be within ±1.87 of `\(\mu\)` .smallCode[ ```r mean(samp) ``` ``` ## [1] 98.22903 ``` ] --- ## Standard error - *SE* is calculated using *N*: there's a relationship between the two .codePanel[ ```r tibble(x = 30:500, y = sd(samp)/sqrt(30:500)) %>% ggplot(aes(x, y)) + geom_line(lwd=1, color = second_col) + labs(x = "N", y = "SE") + annotate("text", x = 400, y = 2.5, label = bquote(sigma==.(round(sd(samp), 1))), color = default_col, size = 6) ``` <img src="data:image/png;base64,#slides_files/figure-html/unnamed-chunk-15-1.png" height="400px" />] --- ## Standard error - That is why *larger samples are more reliable*! <img src="data:image/png;base64,#slides_files/figure-html/unnamed-chunk-16-1.png" height="400px" /> --- ## Standard error - Allows us to gauge the _resampling accuracy_ of parameter estimate (<i>e.g.,</i> `\(\hat{\mu}\)`) in sample - The smaller the *SE*, the more confident we can be that the parameter estimate ( `\(\hat{\mu}\)` ) in our sample is close to those in other samples of the same size - We don't particularly care about our specific sample: we care about the population! --- ## The Central Limit Theorem - Sampling distribution of the mean is *approximately normal* - True no matter the shape of the population distribution! - This is the [Central Limit Theorem](https://en.wikipedia.org/wiki/Central_limit_theorem) - *"Central"* as in *"really important"* because, well, it is! --- ## CLT in action .orig-colors[ 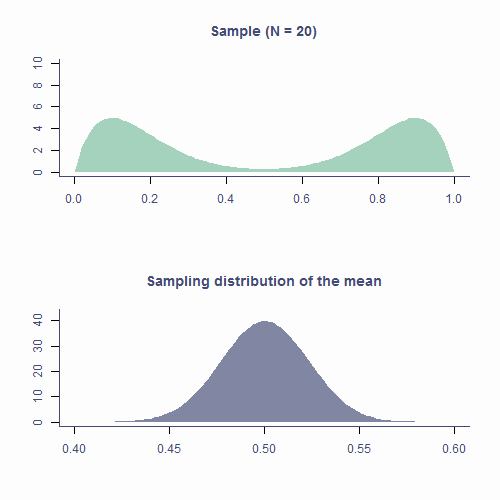 ] --- ## CLT in action .orig-colors[  ] --- ## Approximately normal - As *N* gets larger, the sampling distribution of `\(\bar{x}\)` tends towards a normal distribution with **mean = `\(\mu\)`** and_ `\(SD = \frac{\sigma}{\sqrt{N}}\)` _ .codePanel[ ```r par(mfrow=c(2, 2), mar = c(3.1, 4.1, 3.1, 2.1), cex.main = 2) pop <- tibble(x = runif(100000, 0, 1)) %>% ggplot(aes(x)) + geom_histogram(bins = 25) + geom_vline(aes(xintercept = mean(x)), lwd = 1, color = second_col) + labs(title = "Population", x = "", y = "Frequency") plots <- list() n <- c(5, 30, 1000) ylab <- c("", "Frequency", "") for (i in 1:3) { plot_tib <- tibble(x = replicate(1000, mean(runif(n[i], 0, 1)))) plots[[i]] <- ggplot(plot_tib, aes(x)) + geom_histogram(aes(y = ..density..), bins = 30) + labs(title = bquote(paste( italic(N)==.(n[i]), "; ", italic(SE)==.(round(sd(plot_tib$x), 2)))), x = "", y = ylab[i]) + # xlim(0:1) + stat_density(geom = "line", color = second_col, lwd = 1) } plot_grid(pop, plots[[1]], plots[[2]], plots[[3]]) ``` <img src="data:image/png;base64,#slides_files/figure-html/unnamed-chunk-17-1.png" height="400px" />] --- exclude: ![:live] .pollEv[ <iframe src="https://embed.polleverywhere.com/discourses/IXL29dEI4tgTYG0zH1Sef?controls=none&short_poll=true" width="800px" height="600px"></iframe> ] --- ## Take-home message - *Distribution* is the number of observations per each value of a variable - There are many mathematically well-described distributions - Normal (Gaussian) distribution is one of them - Each has a formula allowing the calculation of the probability of drawing an arbitrary range of values --- ## Take-home message - Normal distribution is - **continuous** - **unimodal** - **symmetrical** - **bell-shaped** - _it's the right proportions that make a distribution normal!_ - In a normal distribution it is true that - **∼68.2%** of the data is within **±1 *SD*** from the mean - **∼95.4%** of the data is within **±2 *SD*** from the mean - **∼99.7%** of the data is within **±3 *SD*** from the mean - Every known distribution has its own _critical values_ --- ## Take-home message - Statistics of random samples differ from parameters of a population - As *N* gets bigger, sample statistics approaches population parameters - Distribution of sample parameters is the **sampling distribution** - **Standard error** of a parameter estimate is the *SD* of its sampling distribution - Provides *margin of error* for estimated parameter - The larger the sample, the less the estimate varies from sample to sample --- ## Take-home message - **Central Limit Theorem** - Really important! - Sampling distribution of the mean tends to normal even if population distribution is not normal - Understanding distributions, sampling distributions, standard errors, and CLT it *most of what you need* to understand all the stats techniques we will cover --- class: last-slide background-image: url("/lectures_assets/end.jpg") background-size: cover
class: slide-zero exclude: ![:live] count: false
---